Details of the objective of the first call for projects of the Priority Research Program on Antibiotic Resistance
What were the objectives of this first call for PPR projects?
Hygiene, prevention and surveillance have helped to reduce antibiotic resistance in several countries. However, it seems that these measures alone cannot completely contain antimicrobial resistance in its entirety. Other alternatives must be supported to control and reduce antibiotic resistance. Whether in human, animal or environmental health, there is a need for research to acquire new knowledge and to understand the host, pathogen and treatment mechanisms that contribute to the emergence of bacterial resistance, its transmission and dissemination in all ecosystems. The knowledge front should make it possible to understand all the underlying mechanisms that make, for example, a bacterial infection resistant to antibiotic treatment, and to elucidate why certain patients, at high risk of infection during hospitalization, do not become infected.
In short, we need to investigate all host mechanisms, including immune, genetic, nutritional and psychological status, which make the host robust or vulnerable to bacterial infection, in order to propose a more effective therapeutic treatment and avoid selective pressure. On the bacterial side, the challenge is to understand all the mechanisms by which bacteria escape from current treatments and alternatives. It is important to know the biology of bacteria in order to find new therapeutic targets, and to understand how multi-resistant bacteria emerge, resist their environment, multiply and persist via reservoirs, and spread to different hosts and the environment.
Support for research must include the development of new molecules, without creating resistance, to avoid therapeutic impasse, as well as new detection tools and early diagnostic tests to stop bacterial colonization as early as possible at host level, at population level (human and animal) to delay possible epidemics, and to control environmental reservoirs. This will make it possible to monitor the global evolution of resistance via standardized, shared and exploitable indicators (notably by taking advantage of the latest technological advances such as artificial intelligence) in all ecosystems. It is also crucial to develop research activity in the human and social sciences, in epidemiology and for interventional studies, in order to describe, analyze and understand the perception of the risk of antibiotic resistance, and to raise awareness among all healthcare professionals and users of the responsible use of antibiotics.
These different fields of investigation – fundamental, clinical, innovative and societal – must be supported at the heart of a single research program, which must also address the challenges posed by antibiotic resistance in countries with limited resources, given the impact of globalization on this issue. An interdisciplinary program bringing together communities of scientists from different backgrounds, some of whom have not yet included antimicrobial resistance in their priorities, would be a real lever for cross-fertilizing skills and expertise to open up unexplored avenues of research and meet the need for innovations, alternatives and technological and behavioral breakthroughs.
The interconnection of disciplines working on a common program would be an asset for boosting research on antibiotic resistance, supporting bold research with controlled risks, broadening the current fields of investigation of academic research and finding opportunities to secure funding to continue the research already underway.
The 4 pillars of the Antibioresistance PPR
The priority research program on antibiotic resistance is based on four interdisciplinary and interconnected pillars:
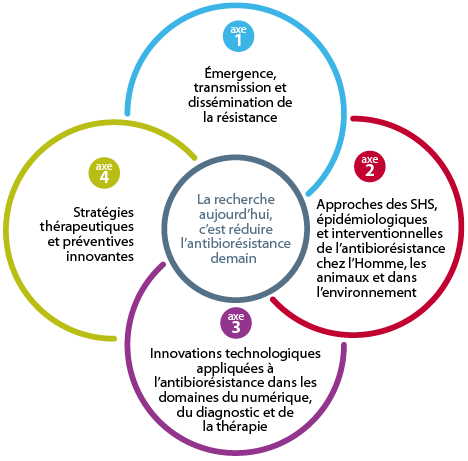
For each axis, the context, issues and research priorities are described, along with an action plan comprising 18 objectives, 53 actions and their indicators. Taken as a whole, the issue of antimicrobial resistance would require a denser program of actions in terms of number of objectives and complexity. It seemed more appropriate to the expert committee to focus solely on antibiotic resistance. To ensure that the RPP remains open-ended and is able to take account of new research and public health issues over the next 10 years, the Scientific Committee, Inserm and its partners recommend that the overall program presented in 2019 should be able to evolve and contribute to supporting challenges that have not yet been identified, depending on the success or otherwise of the challenges that will be undertaken.
Focus 1 • Emergence, transmission and dissemination of resistance
Context
Antibiotics are most often derivatives of natural compounds found in the environment, with their own metabolism. Antibiotic resistance is also a natural process that preceded their use by humans. The use of antibiotics, antiseptics and disinfectants on a bacterial species leads to the selection of resistant mutants that are able to multiply in the presence of these anti-infective agents. The phenomenon of selecting resistant variants to anti-infective agents following exposure exists in microbiology for all organisms, whether bacteria, viruses, fungi or parasites. The same is true of cytotoxic agents used in chemotherapy or insecticides used in vector control. Certain concepts of progressive selection of variants that escape treatment and their dissemination are common to these processes. However, the selection and transmission of bacterial resistance to antibiotics present two particularities that make their dynamics particularly complex. Firstly, these treatments will also affect human and animal microbiota, as well as microbial communities in the environment, by modifying their composition and selecting resistant strains which can then spread to other reservoirs. In addition, an important component of resistance results from the acquisition of resistance genes (ARGs) encoding efflux pumps or enzymes which, for example, inactivate an antibiotic, or modify its target. These resistance genes are frequently carried by plasmid or integrative mobile genetic elements (MGEs), which have the ability to pass from one bacterium to another within the same species, or between species that are even phylogenetically distant. The selection (emergence), transmission and dissemination of antibiotic resistance therefore involve not only the circulation of strains, but also that of EGMs in the multiple reservoirs constituted by human, animal and environmental populations under multiple evolutionary pressures.
Issues
Antibiotic resistance represents a major public health issue for both humans and animals. Surveillance networks exist at multiple levels: national, with Santé publique France for human health, via the CNRs, ONERBA and the national missions of the CPIASs, and ANSES for animal health and the environment; international, with the WHO, OIE and ECDC in particular; but also at local level, at the level of a healthcare establishment. Antibiotic consumption by humans and animals and their release into the environment are also monitored. Alongside these surveillance activities, which are essential for identifying epidemiological changes in time and space, understanding the mechanisms of selection, transmission and dissemination of resistance requires a combination of approaches: i) an evolutionary approach to the selection dynamics of treatment-resistant strains capable of dissemination (and therefore of low biological cost), ii) a molecular approach to understand the mechanisms of acquisition and transfer of resistance genes and mutations, and iii) an ecological approach to understand and model the spatial interactions between microbiota and the intermittent selection pressures to which bacteria are subjected.
A key challenge for the future is to integrate innovative approaches into these studies to gain a detailed understanding of the processes involved (see also axis 3): “omics” approaches (genomics, metagenomics, transcriptomics, proteomics, and also metabolomics), “single cell” or “single molecule” studies, new imaging methods and mathematical and computational approaches, particularly learning approaches.
A second challenge will be to use mathematical tools to combine surveillance data and mechanistic knowledge to model the processes involved in the selection and dissemination of antibiotic-resistant strains and resistance genes in humans, animals and the environment. These models will make it possible to assess and predict the level of risk of acquisition and transmission of antibiotic resistance, associated with local or national antibiotic use policies, preventive healthcare measures, animal husbandry and water treatment procedures.
Research priorities
Selection of resistance genes, resistant strains and resistance mechanisms
The emergence of a resistant clone is a complex process that combines four components that must be studied together:
- Resistance genes and mechanisms: the first step in the selection of a new resistance mechanism is the capture of resistance genes in environmental reservoirs that are often little explored. Paradoxically, the environmental origin of major circulating resistance genes is still unknown. Once captured, resistance genes will have variable potential to evolve to target new antibiotics.
- Chromosomal mutations: the selection of resistant strains also proceeds through the acquisition of mutations (SNPs, indels, recombination, amplification) in various systems directly or indirectly involved in the mode of action of an antibiotic or its accumulation in the bacterium.
- The phenomena of persistence, tolerance and dormancy must be taken into account in the emergence and selection of resistant clones. These phenomena are under the control of genetic determinants that are still largely unknown, and are preliminary stages in the acquisition of mutations conferring resistance.
- The selection and amplification of a resistant strain will depend on the selection pressure exerted by antibiotics and by numerous other molecules and/or non-medicinal practices with a synergistic effect. This collateral selection pressure is often poorly understood, particularly for bacterial populations not directly targeted by treatment (species colonizing microbiota) or in the environment.
Consequently, it is necessary not only to go beyond the catalog of these genes and mutations, but also to integrate the observations into a complex model to reconstruct the dynamics of the appearance of resistant clones and the underlying molecular mechanisms in terms of mutation selection, capture and transfer of resistance genes, and to identify the constraints involved in the success of a resistant clone, such as the compatibility of acquired resistance genes or EGMs with a genetic background, and in the successive acquisition of resistance genes, plasmids and mutations.
Dissemination and transfer of clones and genes – reservoirs
Molecular epidemiology describes, for different species of pathogenic bacteria, the existence of dominant antibiotic-resistant clones and resistance genes associated with EGMs, as well as their distribution in different human, animal and environmental reservoirs. The population size of these clones can vary in time and space, on a local, national or international scale. The reasons for the “success” of these clones are still largely unknown, as are the reasons for their disappearance. Genomic sequencing and international cooperation should make it possible to identify these clones for major pathogenic species, as well as their circulation and geographical distribution in a “One Health” dimension.
The properties of these clones and the genetic determinants involved in their ability to disseminate and transmit between reservoirs are very diverse. They include the ability to colonize and persist in humans and animals or in the environment, resistance to stress, interactions with the host microbiota, and for EGMs carrying resistance genes, their capacity for intra- and inter-species transfer. The acquisition of resistance usually has a biological cost, and the ability to disseminate implies reducing this cost.
Epidemic clones selected by these processes may have particular virulence properties (hyper- or hypo-virulence) which affect the risk associated with these clones and need to be better understood to be taken into account in diagnosis and surveillance. Resistant clones are intermittently subjected to selection pressure by antibiotics or antiseptics, often at low (sub-inhibitory) doses. They are also subject to host defenses (innate immunity, acquired immunity and vaccines) and environmental resilience. It is the result of these evolutionary and ecological constraints, and a clone’s ability to adapt, that determine the dynamics of resistant bacterial populations.
Objectives and action plan
Objective 1 – To map the biodiversity of antibiotic resistance in the three Human-Animal-Environment (HAE) sectors and characterize the intra- and inter-sector transmission routes, as well as the bacterial and environmental factors involved in transmission.
Action 1: We will set up hospital, community, animal health and environmental study sites (based in particular on existing workshop sites) enabling us to develop different research programs in environments monitored over time and space. These studies may include interventional study designs.
Action 2: By interacting with surveillance organizations in the human (Santé publique France), animal (ANSES) and environmental (AFB, INERIS) fields, we will set up methodological research to optimize the collection of surveillance data on antibiotic resistance and antibiotic consumption, as well as the collection and conservation of bacterial strains. The aim will be to better integrate data from the three sectors in order to characterize emergence and transmission pathways.
Action 3: Antibiotic resistance is a global issue, with the worldwide circulation of BMR and resistance genes via various genetic elements. We will encourage collaboration within international networks and analysis of the impact of international trade. Partnerships with low- and middle-income countries will be particularly encouraged.
Action 4: We will create a one-stop shop for biobanks of bacterial strains, particularly BMR, genetic supports and resistance vectors. We will develop “omics” databases (complete bacterial genomes, resistome, metagenomic data from complex matrices – human or animal tissues or biological fluids, environmental matrices – transcriptomic and metabolomic data, etc.) from the three HAE ecosystems, which will be accessible for research purposes, taking into account all confidentiality constraints (in collaboration with axis 3).
Action 5: We will develop quantifiable resistance markers to assess the importance of different routes of antibiotic resistance dissemination (hospitals, direct contacts, international flows of people and consumer goods, the human food chain, farm animal feed, wastewater treatment, manure spreading, wildlife, circulation of biofilms on plastic waste, etc.) within and between HAE ecosystems.
Action 6: We will develop new methods for modeling transmission and emergence, using artificial intelligence-based learning technologies (e.g. machine learning, deep learning) and incorporating the data acquired in Axis 1, Objectives 2 and 3.
Indicators
- Standardized tools accessible to a broad community of scientific and socio-economic players, enabling detailed analysis and comparison of dissemination pathways to go beyond existing surveillance networks (human and animal) and measure the effects of preventive measures.
- Within 5 years, microbiological and omics databases specific to resistance in the three HAE ecosystems will be available for use by the scientific community.
- Mathematical and computational tools to model the emergence, evolution and transmission of antibiotic resistance, and the impact of interventions.
- Modeling the emergence and dissemination of hyperepidemic BMR clones and genes and their capacity for intersectoral transmission.
- Identification of environmental reservoirs of major resistance genes in human and veterinary medicine and exchange dynamics.
Objective 2 – To improve treatments for bacterial infections by identifying bacterial or host-induced escape mechanisms from antibiotic treatments.
Action 7: We will develop relevant in vitro, in vivo & ex vivo study models to highlight specific mechanisms of antibiotic and antiseptic resistance and treatment escape in different environments. These data will be combined with clinical and microbiological data obtained in situations of therapeutic failure.
Action 8: We will investigate the genetic basis of resistance and escape to treatments (biofilm, persistence, tolerance, non-cultivable bacteria, etc.), in order to understand molecular mechanisms, identify new bacterial targets and optimize treatments and their combinations.
Action 9: We will determine the host factors contributing to treatment efficacy: genetic and immune factors, underlying pathologies, accessibility of the infectious site, composition of microbiota.
Indicators
- Validated methods and reference protocols for correlating antibiotic doses and treatment efficacy.
- Identification of new bacterial targets.
- Identification of biomarkers of treatment efficacy and risk of resistance selection for personalized treatment of long-lasting and/or recurrent infections.
Objective 3 – Stop the increase in resistance and reverse the curve.
Action 10: We will define the spatial and temporal dynamics of the acquisition of antibiotic resistance by bacteria, the ecological advantages and modes of transmission of disseminated resistant bacterial clones, resistance genes and genetic carriers of resistance. Multidisciplinary approaches involving clinical and molecular microbiology, microbial ecology, epidemiology, modeling and artificial intelligence will be favored. The effect of antibiotics and biocides in the three HAE sectors, and particularly in hospitals, on the selection and dissemination of resistance will be analyzed to characterize their impact and help design and model interventions (Axis 4).
Action 11: We will identify resistance mechanisms to new antibiotics and their potential for transfer to pathogenic bacteria by screening various samples from the three sectors across different countries. This will allow us to pinpoint high-risk niches and implement preventive measures to limit their spread.
Action 12: We will develop innovative methods to block the transmission of resistance genes. To achieve this, we will characterize the molecular basis of resistance gene capture—particularly from environmental bacteria—their integration into mobile genetic elements (e.g., plasmids, transposons, integrons), and their transmission across the HAE sectors. We will define the interactions between resistance genes, mobile genetic elements, and host bacteria that contribute to their spread.
Action 13: We will develop innovative processes to prevent the dissemination of antibiotics and biocides targeting bacteria into the environment, along with analytical methods to assess their impact.
Indicators:
Recommendations on antibiotic use, as well as infection control and prevention strategies, waste management, or livestock organization, to prevent the transmission, colonization, and spread of high-risk clones.
Innovative “anti-transfer” strategies for resistance genes and environmental decontamination methods targeting antibiotics and biocides.
Axis 2 • Social Sciences, Epidemiological, and Interventional Approaches to Antimicrobial Resistance in Humans, Animals, and the Environment
Context
The use of antibiotics—from production to final use, including prescription and dispensing—varies greatly today, as do the regulatory frameworks designed to control them. It is therefore essential to better understand the underlying logics that lead to antibiotic misuse and to identify dynamics that promote improved prescribing and usage practices. Combating antimicrobial resistance thus requires:
- Analyzing, understanding, and describing contextual determinants and social factors; identifying economic logics, individual or professional practices, legal frameworks, discourses, and situations related to this issue; and observing the populations concerned, decision-making environments, and the arenas where antimicrobial resistance and the proper use of antibiotics are made visible;
- Reducing antibiotic use in human and veterinary medicine to what is strictly necessary, monitoring their use, and promoting prudent/responsible/appropriate usage through Antibiotic Stewardship Programs;
- Combating the transmission of pathogenic bacteria (whether susceptible or resistant) and resistance genes. These actions must be implemented within communities, long-term care facilities, and healthcare institutions. While hospital-based infection control programs address this last point as part of routine activities, their limitations highlight the need for dedicated research projects on this topic.
Challenges
Research objectives should include issues related to social sciences, broad prescribing interventions, and preventive measures. They will focus on understanding and analyzing contexts, practices, and discourses, and will be oriented toward interventions. This analytical and understanding component should be grounded in well-established concepts from the social sciences, though not exclusively. Projects will also focus on antibiotic stewardship and infection control programs related to resistant bacteria, including interventional and quantitative epidemiological approaches. Interventions aimed at addressing antibiotic resistance broadly—covering prescribing, transmission, and both individual and collective impacts—should rely on diverse methodological designs. Lastly, studies incorporating an international and comparative dimension are strongly encouraged.
Research Priorities
Analyze, Describe, Understand
- Knowledge, Information, Communication, and Practices
A key challenge lies in understanding how antimicrobial resistance (AMR) is perceived in different contexts: in hospitals, in human medicine (both primary care and specialist care), and in veterinary medicine (including rural settings and pet care). This includes assessing the level of awareness among stakeholders (healthcare professionals, patients, and pet owners), the availability and circulation of information, as well as the forms and effectiveness of communication about AMR. - Professional and Organizational Dynamics
Antibiotic use is heavily influenced by the professional settings in which it occurs. Hospital restructuring, changes in the agri-food sectors, healthcare working conditions, and the evolution of medical and veterinary professions all play a role. How do these structural changes impact antibiotic prescription and use? Furthermore, preventive measures implemented for patients infected with resistant bacteria in hospital settings have consequences that need to be analyzed in depth.
- Economic and Environmental Issues
AMR sits at the crossroads of multiple competing interests, including the production and marketing of both old and new antibiotics, as well as therapeutic innovations. The economic model of antibiotics is itself a major issue, far beyond direct cost calculations. Additional concerns include how livestock farming and the agri-food industry are responding to changing consumer demands. Finally, the issue of industrial, hospital, and agricultural waste discharge is critical. - Public Policy and Regulation
How AMR has become a public issue, and how that has shaped the implementation of public policies, varies by country depending on its history, culture, and stakeholders. Evaluating the impact of these policies raises the question of which measures are best suited to sustainably reduce antibiotic consumption and infections caused by resistant bacteria.
Control and Prevention of the Spread of Antibiotic-Resistant Bacteria
The emergence and spread of antibiotic resistance is a major public health issue, as evidenced by the increasing incidence of infections and colonization. Hospitalized patients, especially in intensive care units, are highly exposed to healthcare-associated infections caused by emerging highly resistant bacteria (eHRBs). For example, the mortality rate for bloodstream infections in these units ranges from 25% to 30%.
eHRBs—including carbapenemase-producing Enterobacteriaceae (CPE) and vancomycin-resistant Enterococcus faecium (VRE)—are commensal gut bacteria with a high transmission potential, representing a major public health concern both nationally and globally, as recognized by the WHO and other scientific organizations. In France, 2,385 CPE episodes were reported to the Institute for Public Health Surveillance between 2004 and December 2015. Klebsiella pneumoniae is the most frequently isolated species, yet fewer than 1% of its strains are carbapenem-resistant. In contrast, resistance levels in countries like Greece and Italy reach 61.9% and 33.5%, respectively.
In France, rapid detection of eHRBs and early implementation of preventive measures are recommended by the French High Council for Public Health (HCSP) and form a key component of the National Program for the Prevention of Healthcare-Associated Infections (Propias).
These preventive measures are restrictive for both infected or colonized patients and healthcare facilities. They include screening all “contact” patients (i.e., those cared for by the same team before the index case was isolated), and may lead to admission and transfer suspensions. Patient isolation, while crucial for limiting cross-transmission, has psychological impacts, reduces healthcare provider visits, and increases the risk of undetected adverse events. Transfer delays and refusals are also common. The impact on other patients in the same unit is still unknown. Nearby healthcare facilities may also be affected. Moreover, the current activity-based hospital funding system (T2A) depends on a high patient turnover rate. Any disruption to care flow results in immediate financial losses. Thus, in the case of an outbreak alert, two opposing paradigms emerge: limiting admissions and transfers is advisable but leads to decreased funding.
Acting: Intervening
- Observational Studies
Develop large-scale, routine-use measurement tools and indicators to assess the effectiveness of interventions aimed at improving antibiotic practices and reducing epidemic risk. For example, the ability to routinely and automatically measure the quality of antibiotic use in France through national health insurance reimbursement data.
- Interventional Studies
The goal of these interventional studies is to improve practices (through antibiotic stewardship programs or infection control programs) in order to ultimately reduce antimicrobial resistance. They must rely on high-level evidence designs, such as cluster randomized or quasi-experimental designs (with control groups and interrupted time-series analysis). Evaluating barriers and facilitators, as well as conducting a process evaluation (aligned with the methodology of complex interventions), is strongly recommended. Primary outcomes should have clinical relevance and include a quality-of-practice dimension. Other potential outcomes may include:- Medical-economic impact
- Missed opportunities for care
- Adverse event rates
- Medicolegal consequences
- Educational impact
- These interventions (targeting the general population, hospitalized patients, veterinary medicine, prescribers, the agricultural sector, media, and other stakeholders) will require well-defined outcome criteria. Strong emphasis will be placed on cost-effectiveness indicators. Interventions may be implemented across various contexts, including collaborative studies with emerging countries.
- Antibiotic Stewardship
Antibiotic stewardship is a comprehensive strategy aimed at ensuring and promoting the responsible and appropriate use of antibiotics. While commonly translated into French as programmes de bon usage des antibiotiques, the English term encompasses a broader scope (https://www.ncbi.nlm.nih.gov/pubmed/28882725). - Antibiotic stewardship takes a systemic approach, addressing:
- The overall organization of society
- The structure of the target system (human/animal health, environment)
- Professionals in the targeted sector (both prescribers and non-prescribers)
- Antibiotic users (patients in human medicine, farmers in veterinary medicine)
- The general public
- It applies to human medicine, veterinary medicine, and environmental antibiotic use (e.g., in agriculture), within a One Health perspective.
- Control and Prevention of the Spread of Antibiotic-Resistant Bacteria
- Controlling the spread of antibiotic-resistant bacteria requires appropriate and rigorous measures. These measures are relevant not only in hospital settings but also in other environments. However, many of them are based on evidence with limited robustness and rarely incorporate cost-effectiveness analysis.
- New research projects should assess the impact of implementing such measures, including social and behavioral factors. Both endemic and epidemic contexts should be explored. Well-designed studies, even those yielding negative results, must also be reported.
- Control efforts apply to human medicine, veterinary medicine, and environmental contexts (e.g., agriculture, wastewater treatment plants), all under a One Health framework. As stated in the section “Analyze, Describe, Understand,” interventions should clearly assess the impact of antibiotic resistance and prevention measures on:
- Infection control
- Access to care
- Organization of healthcare services
- Lost opportunities for patients, whether or not they are carriers of resistant bacteria
Objectives and Action Plan
General Principle
Issues related to the Social Sciences, Public Health, and Epidemiology revolve around three main objectives:
- Establish a network for analyzing discourses, practices, and uses related to antibiotics and antimicrobial resistance, with an initial focus on: the socio-economic system, the cultural and environmental context, as well as the socio-technical systems in both human and animal domains that help to understand the socio-cultural and contextual roots of antimicrobial resistance. This also includes the “behaviors” of stakeholders, and especially the interactions between caregiver and patient, veterinarian and animal, and farmer and animal that lead to prescription.
- Evaluate control and prevention measures on the spread of antibiotic-resistant and emerging highly resistant bacteria, as well as potential adverse effects at both individual and organizational levels.
- Develop interventional studies in the field of Antibiotic Stewardship to ensure and promote the responsible and appropriate use of antibiotics.
Objective 1 • Establish a network for analyzing discourses, practices, and uses related to antibiotics and antimicrobial resistance within a One Health perspective.
The goal is to analyze, understand, and describe contextual factors and socio-cultural conditions, to identify economic logics, individual or professional practices, situations, legal frameworks, discourses, and language related to antibiotic use and the development of bacterial resistance. It also involves observing the social groups concerned, decision-making environments, and the arenas where antimicrobial resistance becomes visible, as well as placing practices and usage into historical perspective.
This research requires the networking of scholars from various disciplines within the social sciences (sociology, political science, information and communication sciences, linguistics, anthropology, economics, history, geography, etc.), who can collaborate with researchers in human, animal, and environmental health to create pools of expertise capable of addressing all aspects of the antimicrobial resistance issue.
In addition, this observatory could serve as a resource for managing potential crises—whether related to monitoring alert thresholds, informing and communicating with the public, or engaging with public or private decision-makers.
The primary objective is therefore to build an academic community whose research activities can be organized around the following four thematic areas.
Action 14: We will study the dissemination and circulation of information, knowledge, and communication. Communication about antimicrobial resistance raises numerous questions that touch on the very definition of communication itself and challenge the nature of the information, knowledge, and understanding it makes visible and public.
We will examine both professional and lay knowledge that guide antibiotic use in human and animal health, as well as awareness of its environmental impact across different cultural contexts. Attention will be paid to interpretative frameworks and to the concepts (such as “One Health”) that shape how knowledge, information, and representations are made public.
Researchers will focus on actions, discourses, and imagery, as well as the roles played by the media and social networks. The processing and production of digital data also fit into a broader dynamic of knowledge generation and circulation that warrants close attention.
Indicators
- Increased communication and expansion of communication spaces
- Improvement of messaging and information campaigns (empowerment)
- Rise in public awareness
Action 15: We will study practices, work, and organizations. It is essential to consider the impact of professional and organizational contexts on antibiotic use. Restructuring in hospitals and agri-food chains affects the practices of healthcare workers and livestock professionals, transforming their roles and working methods, and ultimately reshaping their use of antibiotics and/or their alternatives.
How are medical (urban and hospital-based) and veterinary professions evolving, and what impact do these changes have on antibiotic use? What types of knowledge are mobilized in the use of antibiotics and their alternatives? Which socio-technical systems and organizational models (e.g., farming systems, types of medical and veterinary practices) promote rational use? What kinds of interactions and contractual relationships between healthcare professionals and their patients/clients are most effective in regulating antibiotic prescriptions?
This action will also examine the use and effects of new digital tools in these contexts, whether for diagnostics or surveillance.
Indicators
- Reduce the number of prescriptions
- Implement preventive and alternative measures
- Integrate these data into the reorganization of healthcare systems
Action 16: We will analyze the economic and ecological challenges. The development and use of antibiotics, as well as their alternatives, take place within economic and market contexts that may either encourage or limit their use.
In human health, the strategies of pharmaceutical industries and questions related to national health insurance coverage for antibiotic alternatives are key concerns. In animal health, the evolving veterinary drug market (production, distribution, sales, etc.) requires close observation. In environmental contexts, the ecological impact of waste discharges (industrial, hospital, agricultural, etc.) must be assessed in terms of both ecological and financial cost, as well as the impact of climate change on the global phenomenon of bacterial resistance to antibiotics.
These studies must support the economic and ecological transition of agri-food systems and the implementation of sustainable antibiotic use.
Likewise, in the food sector, we will study the development of labels and specific quality marks for “antibiotic-free” products, which contribute to changing practices—practices that are also influenced by consumer demand and the strategic positioning of private-sector players (agri-food industries, retail, catering, etc.).
More broadly, studies are also expected on the economic impact and cost of antimicrobial resistance, along with a more general reflection on the economics of antibiotics and its unique features (challenges, market and economic models).
Indicators
- Promotion of sustainable use of antibiotics and their alternatives
- Increase in incentives (to stimulate R&D, the prescription of alternatives, and antibiotic-free production methods) to support the ecological transition of agri-food systems
- Reduction in usage and antimicrobial resistance (AMR)
- Reduction in the overall economic and ecological impact of AMR
Action 17: We will study forms of public action and regulation. It is important to understand the frameworks, as well as the historical and cultural roots of public policies aimed at reducing antibiotic use, and to evaluate their impact.
How has the issue of antimicrobial resistance emerged in public debate? What role have media and communication played in these processes? Who are the actors, and what are the tools of public action in the fight against antimicrobial resistance? What are the effects of these public policies, and which measures appear best suited to promoting a sustainable reduction in antibiotic use and infections caused by resistant bacteria? What ethical issues do they raise?
A reflection on the legal dimensions of the issue is also expected (legislation on prescription/dispensation, antibiotic therapy counseling, etc.).
Indicators
- Implementation of public policies adapted to contemporary socio-economic and cultural changes
- Development of innovative communication and regulatory tools
- Reduction in antibiotic use and AMR
Objective 2 • Evaluate control and prevention measures on the spread of antibiotic-resistant and emerging highly resistant bacteria, and the potential adverse effects at both individual and organizational levels, including in veterinary medicine.
The objective is to confirm the effectiveness of recommended preventive measures or to identify new preventive approaches, to explore the potential impact of these infection control measures on a patient’s loss of opportunity when colonized or infected with antibiotic-resistant bacteria, and to document any disruption in healthcare service delivery at the affected unit level.
Action 18: We will implement interventional studies aimed at combating the transmission of pathogenic bacteria (whether susceptible or resistant) and resistance genes, and improving epidemiological surveillance. These actions will be tailored to:
- Human health: in the community, long-term care facilities, and healthcare institutions
- Animal health: in farms, animal transportation circuits, slaughterhouses or first collection points for animal products, and veterinary practices
- Environmental health: in wastewater treatment plants and effluent spreading areas
These studies will be part of infection control programs for human health and should preferably use experimental or quasi-experimental designs. In hospital settings, they should measure the reduction of cross-transmission of MDR (Multi-Drug Resistant) or eHRB (emerging Highly Resistant Bacteria) due to the implementation of appropriate preventive measures (e.g., contact precautions, isolation, cohorting), and assess the feasibility and impact of innovative prevention measures in different care contexts, such as pediatrics or nursing homes.
We aim to propose innovative epidemiological surveillance strategies (linked to Axis 1), capable of describing the continuum between healthcare institutions and community care regarding MDR/eHRB. In hospitals, dedicated digital tools should be developed to integrate clinical, microbiological, pharmacy, and patient mobility/transfer data, allowing not only early alert detection but also the routine management of MDR/eHRB cases (index cases, secondary cases during outbreaks, etc.).
To a lesser extent, these tools could also contribute to regional or national surveillance networks, helping to describe and anticipate epidemiological trends or threats. These systems will be based on the Health Data Hub, a national infrastructure under development, intended to extract and link data from hospitals, outpatient care, and laboratory results.
In the long term, new surveillance methodologies could be integrated into routine use through national public health agencies.
A cost-benefit analysis will help generate evidence-based recommendations, including health-economic evaluations of screening strategies for MDR/eHRB (e.g., index case and contact screening).
Some of these actions are intended to be implemented in various geographical contexts, including international collaborations.
In animal health, innovative surveillance and prevention strategies will be proposed, distinguishing between companion animals and livestock, while also considering animal movement circuits and the potential for bacterial resistance gene exchange between microbiomes.
In environmental health, similar distinctions will be made between urban, industrial, and agricultural effluents.
Indicators
- Reduction in cases of transmission/secondary cases of MDR/eHRB patients within hospitals and veterinary facilities; implementation of appropriate digital surveillance tools and pilot studies; estimation of direct costs for MDR/eHRB carriers (and controls).
- Sustained reduction in transmission.
- Validated detection capability for endemic and epidemic data, including:
- Description of the “interactions” between community, healthcare institutions, veterinary, and environmental sectors regarding MDR/eHRB (e.g., community strains appearing in hospitals and vice versa);
- Cost-benefit analysis results;
- Identification of factors that facilitate or hinder the spread of resistance and its transfer between different human, animal, and environmental microbiomes.
- Confirmed reduction in spread; proposal of recommendations for hospitals and outpatient care, veterinary medicine, and effluent management across various sources; flexibility of the surveillance system to adapt to emerging epidemiological trends.
Action 19: Estimate the impact of managing MDR/eHRB hospital episodes (isolated or epidemic) on the risk of loss of opportunity for carriers, contacts, and other patients.
Indicators
- Count the number (or proportion) of situations involving a loss of opportunity, and describe the types of loss of opportunity.
- Number and types of loss-of-opportunity events over time.
- Reduction in the number of loss-of-opportunity cases.
Action 20: Estimate, in aggregate, the impact of managing MDR/eHRB episodes at both the care unit and facility levels on access to care for carriers, contacts, and other patients in affected departments. This evaluation will include the analysis of delays in patient transfer/movement, delays in hospitalization, cancelled admissions, and inappropriate early discharges.
The care burden must also be quantified to calculate the hospital resources consumed in these situations, which can then be compared to scenarios without MDR/eHRB carriers. If available, activity metrics will supplement this analysis. These actions will support anticipating the consequences of future alerts and adapting the healthcare system to emerging MDR threats.
Indicators
- Impact of MDR/eHRB case management on healthcare organization; quantification of hospital resources consumed
- Adaptation of service management and healthcare organization; maintaining expected activity levels or managing the impact of control measures on healthcare delivery
- Recommendations for patient care protocols that control MDR/eHRB spread without compromising healthcare delivery, regardless of patient type (non-carriers, contacts, index cases) or population (pediatric, adult, elderly)
Objective 3 • Develop interventional studies in the field of Antibiotic Stewardship to ensure and promote the responsible and appropriate use of antibiotics.
The intervention refers to a uni- or multimodal strategy (e.g., audit and feedback, education, reorganization of care delivery) aimed at improving the quality of antibiotic use (prescription, dispensing, utilization). The focus here is on the “HOW” of Antibiotic Stewardship—meaning strategies designed to improve the quality of antibiotic therapy (e.g., education)—and not the “WHAT”, which refers to defining what constitutes appropriate antibiotic therapy (e.g., determining the optimal duration of antibiotic treatment in terms of effectiveness and resistance risk) (see PMID: 28750920).
Studying the determinants of antibiotic prescription/dispensing/use (as outlined in Objective 1) is a valuable prerequisite for tailoring the interventional strategy as effectively as possible.
Action 21:
We will implement interventional studies aimed at reducing antibiotic use in human and veterinary medicine to what is strictly necessary, monitoring their use, and promoting prudent/responsible/appropriate use (Antibiotic Stewardship Programs). These interventional studies are designed to improve practices through stewardship programs, with the ultimate goal of reducing antimicrobial resistance.
Interventions must employ high-level evidence designs, such as cluster randomized trials or quasi-experimental designs (with control groups and interrupted time series analysis). An evaluation of barriers and facilitators, along with a process evaluation (see complex intervention methodology), is strongly recommended. Primary outcomes must be clinically relevant (see PMID: 29133158).
Establishing a multidisciplinary scientific committee (e.g., researchers from health and social science fields) is highly recommended.
These interventions may target multiple audiences: the general population, hospitalized patients, veterinary medicine, prescribers, the agricultural sector, the media, and other stakeholders. In both human and veterinary medicine, all care settings (healthcare institutions, long-term care facilities, outpatient care) and specialties (general practice, other medical specialties) are included.
Indicators
- Number of funded interventional studies on Antibiotic Stewardship
- Number of innovative interventions validated through these funded studies, with a high level of evidence (see EPOC criteria)
- Reduction in antibiotic consumption and increase in the prevalence of appropriate antibiotic therapies in France following the implementation of these innovative interventions
Axis 3 • Technological Innovations Applied to Antimicrobial Resistance in the Fields of Digital Health, Diagnostics, and Therapy
Context
Technological innovation holds a central position in the health sector and will be addressed in this plan through three main areas: big data and artificial intelligence, diagnostics, and therapeutic innovation.
There is currently no “off-the-shelf” solution to address the AMR challenge, and massive research efforts are required. This field remains underexplored in terms of technological innovation, despite the urgent need and expectation for breakthrough advancements.
In the diagnostic domain, innovations—closely linked to Axes 1 and 2—will support the rational use of antibiotics, enable faster and improved patient management, facilitate the isolation of patients carrying multidrug-resistant bacteria in hospitals (to prevent their spread), allow the identification of new resistance mechanisms, and enhance surveillance of resistance propagation.
In the therapeutic domain, in connection with Axis 4, such innovations will enable the development of new molecules or combinations, as well as the repurposing of older, previously ineffective drugs. They will also support the combination of antibiotic therapy with alternative strategies in both human and veterinary medicine.
Challenges
Big Data and Artificial Intelligence
The use of observational, real-world data—routinely produced in an increasingly systematic and standardized way by the healthcare system—should make it possible to develop new surveillance tools offering both current and forward-looking views of antibiotic resistance. Among other things, these tools will enable the tracking of resistant bacterial strains.
These tools will also be essential for understanding the actual use of medications and the pathologies for which they are prescribed, in both human medicine (primary care and hospitals), especially using the French National Health Data System (SNDS), and in veterinary medicine, as well as for evaluating antibiotic discharge into the environment.
In connection with fieldwork, machine learning methods should help guide the most appropriate use of antibiotics, for example by assisting in the interpretation of diagnostic test results and supporting decision-making algorithms for prescribing, thereby reducing unnecessary antibiotic use.
In this same perspective—and in alignment with Axis 1—the application of recently developed methods should enable progress toward personalized medicine, through the use of genomic data from patients and the bacteria affecting them.
More broadly, these methods should enhance the analysis and more efficient use of the massive datasets generated across the other axes.
Innovation in Diagnostics
A major challenge lies in developing tests that can quickly determine the type of infection, the bacteria involved, and the nature of antibiotic resistance.
To limit inappropriate antibiotic prescriptions and choose the right treatment, it is essential to rapidly distinguish between bacterial and non-bacterial infections. A test already exists for identifying streptococcal throat infections in medical offices and has proven useful in rationalizing antibiotic prescriptions. Now, these applications must be expanded to other diseases and biological samples, identifying specific biomarkers in the process.
For confirmed bacterial infections, bacterial species are currently identified only after a culture step, most often followed by MALDI-TOF mass spectrometry. Identifying antibiotic resistance still requires another culture-based susceptibility testing step. Faster tests that can identify the species and its antibiotic susceptibility—while shortening or eliminating the culture step—must be developed. The clinical impact of such tests will also need to be evaluated.
These diagnostic tools should be adapted to “field use” and “Point-of-Care/Point-of-Need” settings, aligning with the WHO’s ASSURED criteria (Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free, and Deliverable) for use in both low-income countries (with limited infrastructure) and high-income countries (primary care settings).
In parallel, the development of high-tech tests will provide greater precision, supporting the identification of new biomarkers and emerging resistance.
Integrating disruptive technologies—such as microfluidics, digital tools, biosensors, and synthetic biology—into diagnostic development will enhance sensitivity and ideally enable testing directly on clinical samples.
These tests must be sensitive and specific, high-performing and robust, with high predictive values, and suitable for integration into the medical decision-making process.
Therapeutic Innovation
Therapeutic innovation faces several key challenges:
- Developing a robust pipeline of effective new molecules,
- Ensuring these molecules can enter clinical trials with adequate financial support.
An active upstream interdisciplinary research effort is essential to accelerate preclinical studies.
At the same time, the constraints specific to antibiotic development and the unique dynamics of the antibiotic market must inform strategic investment decisions to ensure clinical trial funding. This includes promoting public-private co-development agreements, inspired by European initiatives like the IMI (Innovative Medicines Initiative).
It is also crucial to support formulation research (galenics), aimed at providing optimal delivery methods for targeting specific organs. Such research can repurpose older antibiotics that had become ineffective.
Support should also be directed toward PK/PD research (pharmacokinetics/pharmacodynamics) to optimize dosing—both for antibiotics used alone and in combination—to improve the benefit-risk ratio and minimize resistance development.
This area of therapeutic innovation is closely linked to Axis 4, which focuses more on applied research. Rational and optimized use of new antibiotics—derived from the discovery of new drug targets (as outlined in Axis 1)—represents one of the key therapeutic strategies to counter antimicrobial resistance (Axis 4).
Research Priorities
In Big Data and Artificial Intelligence
- Hospital Data
- Hospital data are highly detailed but historically poorly structured. The French National Authority for Health (HAS) has developed national quantitative indicators related to healthcare-associated infections (HAIs), but these indicators are difficult to obtain and mostly require extensive chart review, even though their calculation could be automated. This requires an electronic health record containing clinical, biological, and therapeutic data, and more and more hospitals are moving in this direction by implementing data warehouses. However, significant data integration work is needed for large-scale use. This enables a detailed understanding of actual antibiotic use, clinical context, potentially the responsible strains and their antibiotic susceptibility, treatment evolution, the implementation of tools to ensure proper medication use, and finally, the development of automated indicators.
- Community Data
In outpatient care, the variety of practice management software and the dispersion of information across individual medical practices or diagnostic laboratories is a barrier to obtaining the same level of insight.
- SNDS and Health Data Hub Data
Reimbursement data for outpatient and hospital services, as well as hospital diagnoses, are collected comprehensively and in a linked manner within the SNDS. Due to their coverage and volume, this information represents an unparalleled data source in Europe. However, as these data are produced for administrative purposes, they remain incomplete (no indication for prescriptions in outpatient care, no pharmaceutical data in hospitals) and are dependent on evolving administrative rules and coding strategies. The establishment of the national Health Data Hub infrastructure, along with accompanying regulatory changes, is expected to enable the linkage of all data produced in the context of care reimbursed by national health insurance. Many studies will thus be able to rely on this new framework to access the context of prescription, administered treatments, the pathogen involved, and its possible resistance profiles.
- Animal Data
In the animal sector, legislative and regulatory measures adopted under the first Ecoantibio plan and the Agricultural, Food and Forestry Law of October 13, 2014, require many stakeholders in the veterinary drug and medicated feed chain to report the antibiotics they distribute. As in other healthcare sectors, this centralization faces operational challenges commonly encountered at various stages of implementation (data collection, harmonization, transmission, storage, and accessibility). Proper use of these data—both within the animal sector and from an intersectoral perspective—is a key issue in managing veterinary antibiotic prescriptions.This data usage should also allow comparison with antimicrobial resistance surveillance data collected in the animal sector, as well as in human health and the environment, in order to better identify major pathways for resistance selection, including cross-resistance.In this context, the French network RESAPATH, unique in Europe, collects antimicrobial resistance data and provides valuable information for understanding the mechanisms of selection and transmission of multidrug-resistant bacteria (including geographic, zootechnical, molecular, and genomic data). However, data completeness and interoperability with analogous data from other sectors remain areas for improvement.These same observations apply to data collected at the European level by Member States (including France) as part of regulatory AMR surveillance plans at slaughterhouses and in certain animal-derived foods.Ultimately, the veterinary sector’s contribution to the establishment of a global data repository is a major issue, which also intersects with priorities identified in Axis 1.
- Environmental Data
- Environmental research and surveillance networks currently collect data to monitor the circulation of MDR bacteria, genetic elements, and resistance vectors across various components of the environment—from livestock effluents to wastewater, including wildlife and soil. This information varies in nature (bacterial strains, genes, genetic elements, etc.) and originates from different sources (water, soil, wildlife, etc.), which must be precisely described to ensure data usability.Some of these data are already deposited in open-access databases such as GenBank or the Antibiotic Resistance Genes Database (ARDB). However, the creation of a unified database—standardizing characterization methods and enabling operational linkage with data from farms, hospitals, and primary care—is essential to advance our understanding of antimicrobial resistance transmission pathways across different compartments.The development of such a database and the means to exploit it should be aligned with Action 4 of Axis 1.
- Interactions with Other Axes
- These interactions can be numerous, and the deployment of software tools for big data analysis must empower fundamental research. In particular, the priorities of Axes 1 and 3 require the integration of innovative approaches to gain a detailed understanding of the processes involved: “omics” approaches (genomics, metagenomics, transcriptomics, proteomics, metaproteomics, and metabolomics), single-cell studies, new imaging methods, and mathematical and computational approaches, particularly those involving machine learning. Mathematical tools should be used to combine surveillance data with mechanistic knowledge in order to model the processes involved in the selection and dissemination of antibiotic-resistant strains in humans, animals, and the environment. These models will enable the evaluation and prediction of the risk of acquisition and transmission of antibiotic resistance in relation to antibiotic use policies (local or national), healthcare-associated prevention measures, farming procedures, and wastewater treatment practices. As part of Axis 2, epidemiological analysis for causal inference—to assess the effect of an actual or pseudo-intervention using observational data—relies on high-dimensional data adjustment models drawn from the latest advances in artificial intelligence. These methods aim to make the most of the high volume and complexity of the data. They must be applied with particular attention to transparency and reproducibility of the chosen methodology.
In Diagnostic Testing
- For Therapeutic Management
- For therapeutic management to be as suitable and timely as possible, it is necessary to have rapid tests (≤2-4 hours from sampling), inexpensive, easy to use and implement, that can quickly detect and identify the type of infection, the bacterial species, and the relevant resistances. These tests should be able to be conducted directly from environmental or clinical samples (blood, urine, rectal swabs, milk, etc.) in a One Health context.
- For Epidemiological Studies (linked to Axis 1)
The tests must be highly specific and accurate (global and targeted -omic approaches: metabolomics, proteomics, which allow for the precise identification of protein variants/mutants, metabolites, and specific reaction products related to resistance phenomena), capable of being implemented on a large scale, but not necessarily requiring to be rapid or highly sensitive (analysis can be performed from isolated colonies).
- For the Discovery and Validation of New Biomarkers
The identification of new biomarkers for infection and resistance mechanisms will enable the development of more efficient and predictive diagnostic tests.
For these tests, technological developments are necessary at each of the following three stages:
- Pre-analytical, from sample collection to preparation for analysis,
- Analytical, from sample preparation to test execution.
Research should focus on the speed and simplicity of sample preparation for a wide variety and complexity of matrices (biological—human or veterinary—such as blood, feces, urine, sputum, milk, etc., or environmental/food), and for bacterial quantities in the sample that vary greatly (from a few bacteria to 10^6 to 10^8 bacteria/ml or /g). - Post-analytical, from obtaining the result to communicating and interpreting the results. These must be reliable, objective, available on-site or transmitted (connected diagnosis) for potentially remote analysis, as close as possible to the prescriber, and ideally providing an unambiguous and highly personalized therapeutic recommendation. Algorithms integrating test results with all available or potentially accessible data for patients, individually or collectively, could profoundly transform antibiotic therapy practices and become a key component of a renewed Stewardship.
For all types of tests, two major technological categories should be explored without prior assumptions:
- Molecular tests: based on the detection and identification of antibiotic resistance genes, using targeted approaches (PCR) or non-targeted approaches (high-throughput sequencing). The main challenges for these tests lie in reducing their costs, adapting them for field use, and for non-targeted approaches, providing rapid results.
- Phenotypic tests: based on the direct or indirect detection of infection or resistance markers (biomarkers produced by the host or biomarkers produced by the pathogens themselves), their activity, or their consequences (e.g., hydrolysis products of antibiotics, analysis of target modifications, etc.).
All of these approaches are complementary, and it is important to develop integrated approaches that combine different techniques to provide varying levels of information. The challenges include, among others, increasing the sensitivity of these tests, simplifying their implementation, reducing their execution time, and lowering their costs.
In Therapeutic Innovation
- Searching for and Developing New Active Ingredients The identification of active ingredients that act through previously unexplored mechanisms is the cornerstone of developing new antibiotics in the long term. Several molecules, including a monoclonal antibody, are currently in various stages of development. The key factors for success in this search for new drug candidates are as follows:
- Understanding the mechanisms of resistance and bacterial adaptation to their environment (host cell or tissue), as explored in Axis 1.
- Revisiting compound libraries (such as the national compound library, which includes many natural substances) and microbial collections, which may include non-culturable microorganisms (allowing for the exploration of new chemical spaces that can be screened) through a dual approach combining phenotypic models and pathway-specific models.
- Integrating teams from medicinal chemistry, structural biology, molecular modeling, and biology (e.g., programs like “Chemistry for Medicine” from the FRM are exemplary).
- Initiating early pharmacokinetic and pharmacodynamic (PK-PD) studies both in vitro and in animal models to select the best administration protocols (loading and maintenance doses, dosing intervals, total treatment duration, etc.) for clinical trials (e.g., as done in Axis 4).
- Revisiting Active Ingredients with Marketing Authorization
This may involve a new indication, a new route of administration, a new formulation, or new combinations. Research in this area, which often concerns molecules no longer protected by patents, is typically conducted by academic teams, sometimes in collaboration with startups. Several funding calls, such as “Old Drugs for New Bugs”, have been launched in recent years to revisit old molecules that were abandoned due to toxicity, like colistin, using modern PK/PD concepts and technologies. JPIAMR projects aim to search for synergies by combining multiple antibiotics or by combining an antibiotic with a non-antibiotic molecule, such as terpenoid derivatives from natural sources. The development of ethionamide boosters in anti-tuberculosis therapies also illustrates the possibility of finding elegant solutions by combining old and new chemical entities to overcome bacterial resistance to antibiotics. A new phase of this project, which involves two academic teams, an SME, and a big pharma company, could be advanced through the Innovative Medicines Initiative (IMI). This demonstrates that such approaches should not be underestimated. - Improving Targeting of Active Ingredients
- Administering antibiotics directly to the infection site is sometimes the only way to achieve active concentrations or improve the benefit-risk ratio by optimizing the ratio between local and systemic concentrations. The EU has funded the PneumoNP project to develop nanoparticles containing antibacterial peptides for the treatment of Gram-negative bacterial infections. The treatment of bone and joint infections or pulmonary infections caused by P. aeruginosa producing biofilms are other examples where the main challenge is optimizing the antibiotic’s diffusion at the target site. Developing innovative and suitable formulations is perhaps more important than the search for new molecules. However, antibiotic vectorization is not limited to tissue targeting. The lack of effectiveness of an antibiotic may result from resistance, poor diffusion to the infection site, or low cellular penetration and retention when bacteria are inaccessible due to their intracellular location (e.g., within macrophages). Formulation optimization can then aim to improve the intracellular diffusion of antibiotics through approaches using nanotechnology or better understanding of transport systems, such as siderophores for innovative coupling. Targeting can also focus on delivering antigens in innovative vaccination/preventive approaches.
Objectives and action plan
Objective 1 • Integrate all health databases and m-health (mobile health) through the development of specific tools and AI in hospital and outpatient medicine, as well as in veterinary medicine and environmental data collection, relying on the Health Data Hub (data quantity, annotation quality, linking).
Action 22: We will develop analyses/sharing of associated data in a standardized format for global epidemiological studies. We will integrate data from other sources (environmental, agronomic, veterinary, etc.) into these developments.
Action 23: We will develop m-health (for health services available continuously via mobile devices) applied to antimicrobial resistance: connected tools, decision/prescription support software, and shared, open, and standardized databases.
Action 24: We will support and propose establishing a link with the Health Data Hub and explore the development of a local hub that includes animal and environmental data.
Indicators
- Number of data sources available under common governance
- Number of users utilizing a mobile health service
- Number of articles involving these data sources and passing through the Health Data Hub
- Analysis report on relevance
Objective 2 • Develop tools for data analysis, sharing, and utilization to conduct epidemiological studies for identifying antibiotic resistance, its evolution, and its spread in connection with Axis 1 (biomarkers, “-omic” approaches, data sharing, monitoring), as well as the most efficient healthcare organizations and interventions to reduce antimicrobial resistance.
The objective is to develop targeted and integrated approaches in the field of antibiotic resistance to identify and validate new bacterial resistance biomarkers from various origins (DNA/RNA/exosomes/proteins/metabolites…), as well as efficient interventions, healthcare organization strategies, or combinations of existing treatments.
Action 25: We will ensure the use of existing infrastructures and frameworks, while respecting data security and confidentiality. The French Institute of Bioinformatics (IFB) will be specifically contacted to explore potential collaboration in this area.
Action 26: We will establish data governance, ensuring its alignment with international standards and supporting its implementation, especially in countries without adequate infrastructure.
Indicators
- Identification and use of infrastructures for hosting and processing big data, including biomarkers and omic data, within a secure framework.
Objective 3 • Enable the rapid differentiation of bacterial infections (gram+/gram-) from fungal, viral, or parasitic infections, and identify the involved pathogenic microorganism, to avoid the unjustified administration of antibiotics.
Action 27: We will identify and validate direct or indirect biomarkers capable of differentiating a viral infection from a bacterial infection through targeted or non-targeted approaches. We will develop sensitive, specific, and possibly multiparametric tests that allow for rapid, low-cost point-of-care identification without the need for prior culture, directly on veterinary and human clinical samples (milk, blood, urine, feces, sputum, saliva). We will promote the development of diagnostic tools that allow for real-time evaluation and better characterization of the immune status of patients at high risk of infection (elderly, intensive care, major surgery, etc.).
Indicators
- Availability of rapid, sensitive, simple, and low-cost field tests to differentiate a bacterial infection from other infections directly in biological and environmental samples.
Objective 4 • Develop early and individualized bacterial infection detection tools in livestock to target interventions, reduce the number of animals treated, and reduce antibiotic consumption.
Action 28: We will support the development and implementation of methods based on artificial intelligence for the early detection of bacterial infections in herds through the analysis of signals from embedded electronic sensors or videos. This individual-level detection is a prerequisite for developing more targeted intervention strategies (diagnostic, therapeutic, etc.) that replace treatments for entire groups.
Indicators
- Availability of tools (algorithms) for early disease detection and the development of intervention strategies that limit antibiotic use.
Objective 5 • Develop faster, more sensitive, and less expensive first-line tests “Point of Need, Point of Care” to identify the bacterial strain and potential resistance mechanisms, in order to administer the correct treatment more quickly and avoid the spread of resistance.
Action 29: We will shorten the duration and simplify the first-line resistance identification test (from sample to result) to eliminate culture steps by developing methods that can be directly used on biological samples at the lowest possible costs in a One Health context.
Action 30: We will develop innovative, cost-effective targeted and non-targeted approaches, both genotypic (whole genome sequencing, metagenomics, metatranscriptomics, PCR) and phenotypic (proteomics, metaproteomics, metabolomics, immunological, biochemical), for confirmation tests, microbial population analysis in the One Health context (food, environment, clinical – normal and pathological conditions: microbiota, infections, veterinary) or for epidemiological purposes, and propose new targets (biomarkers) for developing new first-line tests.
Action 31: We will enhance the performance of tests and their multiplexing capabilities (simultaneous detection of multiple resistance biomarkers). We will propose innovative methods to differentiate and quantify various antibiotic resistance mechanisms in clinical isolates and their potential induction in the context of the host and therapeutic treatments.
Action 32: We will develop antibiotic susceptibility tests that are simple, fast (less than 4 hours), low-cost, and, if possible, decentralized.
Indicators
- Availability of rapid, sensitive, simple, and low-cost field tests to identify pathogens, resistance mechanisms, and antibiotic susceptibility directly in biological and environmental samples.
- Identification and validation of new biomarkers for infection and antibiotic resistance.
Objective 6 • Develop simple decontamination tools and surface decontamination control (inert surfaces, intervention tools, hands, etc.).
Action 33: We will develop new surface disinfectants based on new techniques and molecules (e.g., the use of bacteriophages). In parallel, we will encourage research into new disinfectants for medical devices, such as new detergent-disinfectant molecules and new low-temperature sterilization techniques. We will promote the development of quantification techniques for bacterial contamination and the identification of indicator microorganisms of contamination. These developments will be carried out with consideration for environmental safety and are crucial for combating healthcare-associated infections in both human and animal healthcare settings.
Objective 7 • Accelerate R&D for new antibiotics through chemistry to renew the drug arsenal.
Action 34: We will facilitate the gathering and accessibility of commercial and national compound libraries on a single website, providing them for distribution and access at moderate costs. We will also encourage national chemists to make their private libraries available through mechanisms to be defined (e.g., help with plate preparation, purchasing assistance, and price negotiation). Similar to the model proposed by the Vaincre la mucoviscidose association, which finances breeding and distribution of an animal model for cystic fibrosis (mouse strain) to assist researchers, we will pool screening results from research teams on a dedicated platform (within the limits of intellectual property concerns) to avoid redundant experiments and enable better analysis of progress in the programs.
Action 35: We will encourage multidisciplinary approaches, combining teams in medicinal chemistry, structural biology, molecular modeling, and biologists already involved in the upstream phases of R&D for the discovery of new molecules and the characterization of their antimicrobial activity.
Action 36: We will initiate early PK-PD studies in vitro and in vivo using animal models. We will establish a mapping of platforms capable of conducting in vivo efficacy testing, specifying the species and available infectious models based on required protection levels, as well as specific features (e.g., equipment allowing repeated antibiotic nebulization in rodents) or imaging equipment (e.g., bioluminescence measurement devices). This will be in connection with Axis 4.
Indicators
- Patent filings related to the discovery of new substances with antibiotic or adjuvant properties.
- Acquisition of candidate molecules for preclinical and phase 1 clinical trials.
Objective 8 • Optimize the effectiveness and efficiency of already marketed antibiotics.
Action 37: We will revisit already marketed antibiotics, such as colistin, to test their potential effectiveness against resistant strains, both in monotherapy and especially in combinations, in order to increase efficacy, reduce toxicity, and avoid the selection of resistant mutants.
Action 38: We will develop modern PK/PD approaches, inspired by those used in other therapeutic areas, to optimize the dosage of antibiotics with a narrow therapeutic margin (in connection with Axis 4).
Action 39: We will develop new methodologies to optimize the use of antibiotics in combination.
Action 40: We will select the most appropriate route of administration and adapt formulations and/or develop prodrugs to better target the infection site.
Indicators
- Implementation and publication of clinical pharmacokinetic studies of already marketed antibiotics, focusing on new indications or administration modalities, especially within the framework of hospital clinical research programs (PHRC).
- Acquisition of funding and completion of clinical trials in the context of “old drugs for new bugs”.
Axis 4 • Innovative Therapeutic and Preventive Strategies
Context
Despite the need for new antimicrobials for clinical use, only a small number of antibiotics have been brought to market in the past 30 years, and many pharmaceutical companies have left the field. This is because the discovery and development of new antibacterial agents is difficult, and the introduction of such therapeutics potentially raises significant regulatory issues. Currently, efforts are being made to explore new ways to combat antimicrobial resistance outside the antibiotic paradigm. For example, the use of monoclonal antibodies (mAbs) as antibacterial agents is being considered, but it has not been extensively evaluated. Since they do not bind to the same bacterial targets as antibiotics, they could complement antibiotics in the management of difficult-to-treat infections. Other innovative molecules (peptides, bacteriophages, lysins, etc.) could help overcome current antibiotic resistance due to different mechanisms of action, offering therapeutic opportunities for fighting severe bacterial infections. This must also be contextualized with the notable increase in immunodeficiency or at least immuno-modification, not only related to increased life expectancy but also to scientific advances that allow patients once afflicted by diseases deemed fatal to survive. This strengthens the relevance of therapeutics targeting host defense imbalances, both for the patient (immunotherapy) and the pathogen (anti-virulence).
A general challenge in the drug development process is the lack of mechanisms for researchers, drug developers, and clinicians to share data and experiences from the development of drug candidates. This leads to duplication of efforts and, ultimately, inefficiencies in the process.
Challenges
Every citizen is confronted multiple times in their life with infectious issues and has been, and will be, exposed to antimicrobial therapy. They will, therefore, sooner or later, directly or indirectly, face the risks of treatment resistance. Moving away from the “all-antibiotic” approach is undeniably a public health issue and a concern for everyone. Even though many operators are currently working in the field of therapeutic innovation in bacterial infections at the national level, continuing to develop a scientific-economic ecosystem in this field, connected with preclinical and clinical research actors, is essential and should be supported. Although several French SMEs operate in the field of anti-infectives, and Europe remains very strong in the field of real therapeutic innovations in bacterial infectiology, it must be noted that American biotech companies attract a significant portion of the research budgets in the field. It is therefore crucial to help French and European teams stay competitive in the global fight against antimicrobial resistance.
This is precisely why the BEAM alliance was created to unify the European momentum. Even though this creation and the awareness of the necessary support for these companies have allowed several of them to obtain real means for innovation, the landscape remains heterogeneous, with innovations that deserve more support to reach the patient.
Supporting new strategies leading to real therapeutic innovations should effectively connect relatively different worlds with a One Health approach, considering human health, animal health, and environmental health from the outset.
In human health, the majority of antibiotic consumption occurs in outpatient medicine. Therefore, therapeutic innovation should not be reserved for hospital medicine, even though antibiotic resistance is most prevalent there. One of the challenges lies in involving primary care stakeholders in such a dynamic.
In the animal sector, addressing antimicrobial resistance in its One Health dimension means prioritizing the risks for humans related to antibiotic use in animals, considering the interconnections between Human-Animal-Environment in a global ecological dimension, but also the economic disparities at the international level. Due to this context and the respective levels of antibiotic consumption, the main challenges focus on livestock animals producing food for humans, even though companion animals should not be overlooked.
Finally, since antimicrobial resistance is not confined to our borders, far from it, innovations must consider their accessibility for low-income countries and the most disadvantaged populations from the outset.
Component of the challenge imposed on tomorrow’s agriculture: the globalization of economies and agriculture, growing citizen expectations regarding livestock, environmental impacts, and a reduced reliance on “medicinal inputs,” require a shift toward sustainable farming systems that prioritize animal welfare. The control of bacterial infections is part of this evolution. It relies on integrated animal health management strategies that use a broad range of tools, primarily targeting disease prevention, which will not be addressed in this document.
Priorities for Research
Among the various therapeutic innovations and other strategies, some seem more likely to change clinical practices in the near future, with real benefits for patients, animals, and the fight against antimicrobial resistance.
Human Medicine
Immunotherapy is undoubtedly one of the most promising avenues, not only to immunorestorative patients to help them better defend against infections, especially as more and more patients are immuno-compromised, but also to provide specific therapeutic strategies. In this field, monoclonal antibodies appear to be close to reaching the market, with both preventive, curative, and protective approaches. The first clinical trials have already been conducted, with Phase 3 studies underway for the most promising molecules. Although these strategies are mainly driven by the industry, French academic actors are heavily involved in their validation.
Antimicrobial peptides, although studied for many years, have not yet been successfully used clinically due to negative results in early-phase therapeutic trials. However, this therapeutic class presents numerous advantages and could be positioned as an adjuvant to antibiotic therapy rather than as a strict substitute. These peptides could also be used for their antiseptic properties and as immunostimulants. Despite past development failures, these compounds deserve to be supported and tested again in more targeted therapeutic indications. In this translational phase, academic actors play a leading role.
Although the fight against antimicrobial resistance requires drastic reductions in antibiotic use, antibiotics should remain part of our pharmacopoeia, and their conditions of use deserve reconsideration. One of the priority topics in this field is to evaluate the impact of antibiotics/alternatives on commensal microbiota (especially the digestive microbiota) in terms of the emergence and spread of resistance, using standardized methodologies to classify therapeutic solutions by decreasing risk. Furthermore, before promoting prudent/responsible/appropriate use of existing antibiotics (or antibiotic stewardship, see Axis 2), specific research should be conducted and supported to validate new usage protocols in terms of indications, treatment durations, or antibiotic combinations, with adaptations applicable in both healthcare settings and outpatient medicine. The antibiotic/non-antibiotic combinations, with “adjuvant” agents that optimize antibacterial activity or target resistance mechanisms (such as beta-lactamase inhibitors), also seem promising. PK/PD approaches applied during preclinical phases to analyze in vitro or animal model data (see Axis 3) will define the administration protocols that will be evaluated in a clinical context. New evaluation criteria should also be developed to differentiate molecules correctly, not just based on infection-treatment duration (e.g., impact on virulence, interaction with the immune system, impact on microbiota…). One of the peculiarities of antibiotics, compared to other therapeutic classes, is that they share targets across different bacterial species, defining their spectrum of activity. However, “narrow-spectrum” molecules, or even “monopathogen” antibiotics, which are beginning to be tested in clinical phases, could contribute to solving part of the resistance problems. In the antibiotic chapter, identifying new targets to attack bacteria differently, through new molecules or by drug repurposing, is crucial.
Bacteriophages have been used for decades in some countries, particularly in the former Soviet Union (Ukraine, Georgia), without conclusive proof of their efficacy and safety. However, this approach, based on the properties of certain viruses, could complement the therapeutic arsenal against bacteria, particularly in extremely difficult-to-treat pathologies or for patients facing therapeutic dead ends where treatment options are limited. The primary diseases concerned are pulmonary infections and bone-joint infections, with local administration. It is worth noting that European regulations on this type of product are not necessarily in harmony with those of other developed countries. In France, bacteriophages can be used compassionately as magistral preparations, under the responsibility of a pharmacist, and under ANSM supervision. The main barrier to using and evaluating bacteriophages is the quality of the product. Indeed, bacteriophage production requires bacterial culture, and purifying the final product—eliminating bacterial products, especially exotoxins and endotoxins—requires a specific process. Non-genetically modified bacteriophages are not patentable (natural product). Bacteriophage lysins, which are proteins produced during bacteriophage replication, are also interesting therapeutic avenues as they have their own bactericidal effect. Therefore, research on bacteriophages and bacteriophage lysins should be encouraged and supported. Beyond compassionate use, clinical research in the West on bacteriophages is currently limited to a few early-phase trials led by a small number of industrial actors, with the complexity and regulatory issues hindering their development by academics alone.
The microbiota, which hosts billions of bacteria, plays a central role in bacterial homeostasis, disease resistance/susceptibility, and also in the Human-Animal-Environment interconnection, as the primary reservoir for the spread of antibiotic-resistant bacteria. Strategies to “manipulate” the microbiota, such as probiotics, fecal transplants, or selective decolonization, appear highly promising in this context. Specific research efforts must be made in this area, acknowledging that its complexity makes it probably harder to approach than other fields. This will require the involvement of many different experts, especially as the probiotic/microbiota issue lies at the intersection of human health, animal health, environment, and food. Within this broad theme, the impact of vaccinations on microbiota must also be considered. Fecal microbiota, used for curative purposes in human diseases, is considered a medicinal product, according to Article L. 5111-1 of the Public Health Code. In the absence of market authorization, fecal microbiota (like bacteriophages) can be used in the legislative and regulatory framework applicable to magistral and hospital preparations (Article L. 5121-1 of the Public Health Code). Magistral preparations are prepared extemporaneously based on a prescription for a specific patient and must comply with Good Preparation Practices, including the tracking of medications under a pharmacist’s responsibility. Regarding fecal microbiota, a fecal bank must be created, and the samples must be stored for at least 2 years at -80°C. Microbiota research is already in clinical phases, led both by industry and French academic players, with many projects ranging from early and translational stages to later phases.
Although vaccination cannot be considered a true therapeutic innovation and its widespread application to all species of pathogenic bacteria faces the problem of target identification, it remains a central tool in the fight against antimicrobial resistance. Rather than developing new vaccines—which is extremely complex and mostly the domain of large industrial groups—it is important to focus specifically on the impact of antiviral vaccination on antibiotic consumption and therefore indirectly on antimicrobial resistance.
Alongside certain innovative molecules/strategies that are close to clinical application, other more experimental approaches, with potential responses to various stages of the infectious cycle (prevention, treatment, protection), but also more distantly from clinical use, deserve support. In this regard, essential oils exhibit interesting anti-infectious properties for some of them. Other molecules targeting virulence factors, quorum sensing, or biofilms should be further developed and tested for their clinical relevance. Similarly, predatory bacteria (Bdellovibrio bacteriovorus) and other natural products like honey and propolis, known for their anti-infectious properties, should be explored further in specific clinical situations.
Veterinary Medicine
Beyond the general similarity of bacterial infectious mechanisms between humans and animals, specific characteristics of the animal sector must be taken into account to identify the most promising innovative strategies. For example:
- The animal health market, with a much lower turnover than human health, lacks health insurance, and is highly segmented by species or production sector.
- The negative impacts of treatments on commensal microbiota, particularly the digestive microbiota, are more significant in the “benefit-risk” balance of animal antibiotic therapy. When relevant, the impacts on environmental microbiota should also be considered.
- Regulations specific to medications for animals producing food for humans, concerning consumer safety (residue dossier) and environmental impact (ecotoxicology dossier).
Preventive Strategies for Animals
- Vaccination/Immunotherapy/Anti-virulence:
Research and development led by major industrial groups in vaccination are dynamic, especially for profitable markets such as domestic carnivores and major livestock sectors (cattle, pigs, poultry). Immunostimulation research shows promise. In preventive vaccination approaches, new immunological adjuvants will be essential to obtaining more effective vaccines. - Probiotics and microbiota manipulation/fecal transplantation:
Various strategies targeting the microbiota of animals, with its roles in:- Disease resistance/resilience, through probiotics or certain essential oils.
- Carriage and spread of antibiotic-resistant bacteria, through fecal transplantation or selective decolonization targeting certain zoonotic pathogens. These strategies could also act as adjuvants to prevent real-time microbiota impact on antibiotic resistance.
² Here arises the issue of “minor” species/indications, for which there is a lack of investment from industry due to the low profitability of the market. The development of autovaccines compensates for the lack of vaccines in certain sectors.
³ Limiting to interventions on the animal: interventions on the animal’s immediate environment (management of bedding and waste) are more closely connected with the scope of Axis 1.
Preventive Strategies for Animals
It is necessary to distinguish between “therapeutic solutions,” which align with research priorities for humans, and “intervention strategies in farming,” which are specific to the animal sector.
- Therapeutic Solutions Domain
- Research in immunostimulation is promising, as evidenced by the recent market release of a PRR (Pathogen Recognition Receptor) agonist for cattle.
- Research focusing on antibiotics should be supported, aiming to optimize their use, leading to a significant reduction in quantities used. This involves:
- (i) revisiting and optimizing the conditions for using the current therapeutic arsenal (doses, administration schedules, treatment durations),
- (ii) exploring innovative solutions in terms of antibiotic combinations or combinations of antibiotics/non-antibiotics, with “adjuvant” agents that optimize antibacterial activity.
- Similarly to human medicine, PK/PD approaches applied to the modeling of clinical and preclinical data, such as from innovative in vitro models, provide a relevant framework for evaluating and optimizing the tested solutions.
- Bacteriophages also represent a promising area of research for veterinary applications. Given its specific characteristics and the regulatory context of veterinary medicine, this approach could provide solutions for certain pathologies, either alone or more likely in combination/adjuvant with antibiotic therapy.
- Research into non-antibiotic molecules, targeting factors related to pathogenicity and/or reduced sensitivity to antibiotic treatments, is also relevant. These are distinct approaches, targeting virulence factors, quorum sensing, or biofilms.
Intervention Strategies in Livestock
Research in this area aims to reduce the quantity of antibiotics used for treating bacterial infections in livestock. It focuses on the development of technological and methodological tools for early and individualized detection of bacterial infections in farms. The implementation of these tools will enable the development of more targeted and effective therapeutic intervention strategies, replacing the need for group treatments.
These strategies may have a strong connection with Axis 3 (detection/diagnosis/individualization), particularly through the development of rapid diagnostic tools and point-of-need intervention (point-of-care).
This multidisciplinary field of innovation involves experts in electronics (sensors, embedded systems, RFID chips), image analysis (video), and artificial intelligence, applying mathematical/statistical expertise to machine learning.
Objectives and Action Plan
Ambition: “To bring to market/offer prescribers innovative preventive and curative strategies that reduce the prevalence of bacterial resistance (pathogenic flora and commensal microbiota).”
Objective 1: Develop a standardized methodology to evaluate, during the development of innovative curative and preventive strategies for humans and animals, their impact on the emergence and spread of bacterial resistance, taking into account both pathogenic bacteria and commensal microbiota.
Action 41: We will create expert groups tasked with defining reliable judgment criteria to assess during clinical trials the impact of therapeutic interventions on the emergence of bacterial resistance in pathogenic bacteria and commensal microbiota (and environmental microbiota when relevant).
Action 42: We will support the development of a national professional clinical research network operating in the field of bacterial infectious diseases, integrating all its sensitivities, to meet the requirements for participation in clinical trials evaluating new antibacterial anti-infectives.
Action 43: We will foster collaboration between research actors (especially clinical) and regulatory agencies at both national and European levels to develop methodologies and specific criteria for trials evaluating alternative strategies to antibiotics and generally aiming to reduce antimicrobial resistance.
Indicators
- Creation of a specific ANSM methodology (e.g., fast-track designation or breakthrough) for clinical research in bacterial infectious diseases.
- Existence of a French clinical research network operating in the field of bacterial infectious diseases.
Objective 2: Develop and bring to market innovative curative and preventive strategies that enhance the host’s defenses and promote their action.
Action 44: We will foster collaboration between the infectious disease and immunology communities and encourage joint research on the interaction between immunity and resistance through targeted calls for proposals.
Action 45: We will develop immunorestorative therapeutic strategies by creating new molecules that restore immunological homeostasis in contexts of aggression (chemotherapy, shock, polytrauma, etc.). For livestock, stress episodes lead to altered immune responses, for which the development of immunostimulants represents a promising research avenue. This strategy could also apply to domestic carnivores, similar to indications in human medicine, for chronically infected or immunodeficient animals at high risk of bacterial infection.
Action 46: We will encourage the development of new antibacterial and antiviral vaccines that will reduce the use of antibiotics, particularly during but not limited to, winter infection episodes. Vaccination plays a prominent role in preventive strategies in veterinary medicine, particularly for animals living in groups where poly-microbiota is constant. New vaccines must induce a strong immune response in already infected animals. The development of new adjuvants is a crucial step in achieving more effective vaccines.
Action 47: We will encourage demonstrating the efficacy of non-vaccine therapeutic strategies positioned as alternatives to the prophylactic use of antibiotics in livestock. Strategies targeting the microbiota of animals, linked to its roles in resistance/resilience to diseases, such as probiotics or certain essential oils, must be evaluated using validated methodologies.
Indicators
- Launch of Point-Of-Care tools for immuno-evaluation before antibiotic treatment.
- Monitoring of antibiotic consumption in outpatient medicine during viral winter episodes.
Objective 3: Develop and bring to market innovative curative and preventive strategies that target bacteria.
Action 48: We will support the development of monoclonal antibodies targeting bacteria with high resistance potential (Staphylococcus aureus, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Acinetobacter baumanii) for preventive and curative applications in humans in indications where therapeutic needs are not met. Exploration of monoclonal antibodies in animals will be encouraged, particularly for domestic carnivores due to the market context.
Action 49: We will foster the emergence of research projects aimed at developing innovative curative and preventive strategies targeting virulence factors, particularly bacterial toxins.
Action 50: We will encourage the development of combined antibacterial therapies in humans and animals, including antibiotics (AB) and non-antibiotic solutions (non-AB) such as bacteriophages, antimicrobial peptides, physical agents, etc. These solutions (AB-AB, AB/non-AB, non-AB) must be evaluated for their effectiveness, their ability to reduce the consumption of the antibiotics involved, and ultimately the emergence of bacterial resistance. Concerted actions with the relevant regulatory agencies will be initiated to accelerate the availability of the most effective strategies in critical situations (ATU…).
Action 51: We will encourage research specifically aimed at reducing antibiotic treatment durations and narrowing their spectrum in well-defined clinical situations.
Indicators
- Increase in the number of drug candidates in new therapeutic classes targeting bacteria.
- Number of projects submitted in response to calls for proposals in the framework of PHRC (Clinical Research Hospital Programs) in the field of infectious diseases.
Objective 4: Develop innovative solutions targeting the resistance gene reservoir in the digestive microbiota to prevent or correct the selection/amplification of bacterial resistance induced by antibiotic treatments.
Action 52: We will evaluate the co-benefit of fecal self-transplantation in high-risk dysbiotic situations (chemotherapy, prolonged parenteral nutrition, septic shock…) by encouraging research and therapeutic trials in this field. The co-benefit of fecal self-transplantation lies in restoring the microbiota’s two functions: 1) the microbiota as a “health factor” and 2) the microbiota as a “reservoir of resistance.”
Action 53: We will encourage non-microbial therapeutic innovations positioned as “adjuvants” to antibiotic therapy to prevent “real-time” processes of resistance selection/amplification. These strategies could target bacteria in the microbiota (e.g., selective decolonization using bacteriophages…) or target antibiotics (e.g., destruction, capture). For animals, the development of “eco-friendly” antibiotics whose pharmacokinetics prevent their diffusion in distal parts of the digestive tract is part of this approach. In livestock, these same strategies (fecal transplantation, selective decolonization…) can target the healthy carriage of zoonotic pathogens (Salmonella, Campylobacter…).
Indicators
- Development of fecal microbiota transplantation programs in human and animal health.